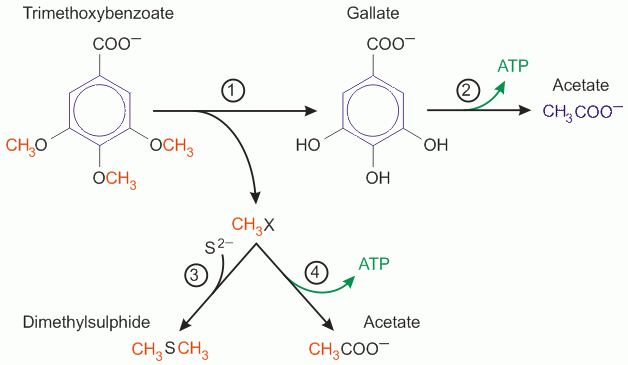
Sketch of the branched catabolism of methylated phenolic plant compounds by Holophaga foetida. Four pathways are involved in the degradation of phenyl methyl ethers: (1) demethylation, (2) aromatic ring degradation by the phloroglucinol pathway,(3) sulphide methylation, and (4) methyl carbonylation by the acetyl-CoA pathway.
Anaerobic Demethylation
During my Diplom and PhD thesis with Bernhard Schink, I worked with a novel homoacetogenic bacterium because of its unusual features. We named it Holophaga foetida since it completely (holos) eats (phagein) its aromatic substrates (both the phenolic ring and the methyl groups of phenyl methyl ethers) and stinks (foetidus) due to the production of dimethyl sulfide from the methyl groups of the phenyl methyl ethers.
Holophaga foetida is a fascinating bacterium that among other things combines the two pathways of demethylation and ring degradation, that were known from its competitors, into one longer pathway, which yields more ATP, but the bacteria grow more slowly than the competitors that only demethylate or only degrade the aromatic ring. Nevertheless, Holophaga foetida is numerically more abundant than the faster growing competitors in all the sediments that we tested. I did not understand why this bacterium grows more slowly during my work in the Schink lab, but many years later while at Bonn, I came across kinetic theory that can in fact explain this observation.