Modelling Salmonella infections of eggs
Human Salmonella Enteritidis infections are mainly due to comsumption of spoiled eggs. There are two ways Salmonella get into eggs, called horizontal and vertical transmission. Horizontal transmission means that the bacteria get through the egg shell into the albumen, which implies that the egg has already been made. Vertical transmission means that the bacteria get into the egg within the chicken while the egg is being made by adding layers around the egg cell called yolk, therefore the bacteria may contaminate the vitelline membrane wrapped around the yolk or the albumen. Anyway, the bacteria will typically find themselves in the albumen more or less close to the honey pot yolk, an environment that has been designed in the course of egg evolution to inhibit the growth of bacteria. Bacteria in the albumen will therefore grow only very slowly, but if they manage to swim to the yolk and enter the yolk through the vitelline membrane, they thrive.
Since eggs become contaminated by low numbers of Salmonella in the albumen, and growth is poor in the albumen but good in the yolk, the question is, how long does it take to reach the yolk?
Watch a movie of a typical simulation.
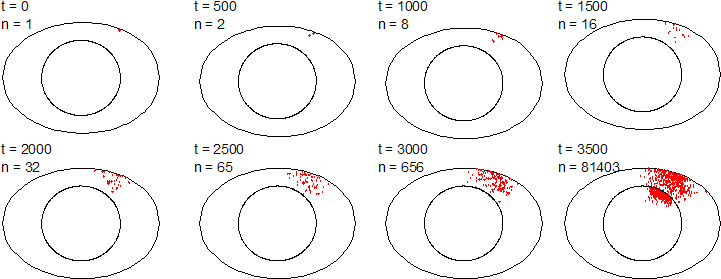
An individual-based modelling approach is well suited to answer this question, because swimming and growth of the initially scarce bacteria is better modelled from the point of view of individual bacteria rather than describing the process on a coarser scale as diffusion of biomass. The more the numbers of Salmonella increase, the better a continous approach becomes. But the egg will be unsafe to eat before this happens. Koen Grijspeerdt from the Centre of Agricultural Research Gent, Belgium, and I have extended BacSim to model the growth and migration of Salmonella in eggs. The image above shows results from a typical simulation.
Motility of the Salmonella bacteria was described by a run and tumble mechanism. For the sake of simplicity, the bacteria were assumed to grow exponentially, an appropriate assumption for the initial phase of growth relevant for shelf-life predictions. Both albumen and yolk were assumed to be homogeneous. The impact of several model parameters (chemotaxis, growth rate, initial contamination numbers and bacterial swimming speed) was assessed by a sensitivity analysis. The results show that chemotaxis towards the yolk would have a strong effect on the time needed to reach the vitelline membrane, an aspect that future research should focus on. The contamination position had less impact on the time to reach the vitelline membrane. The simulation results illustrate the need for more detailed knowledge on the subject of bacterial migration in hens' eggs. Our model can easily incorporate this knowledge when it becomes available. A manuscript describing these results has been accepted by the International Journal of Food Microbiology.
Here is more information on eggs.
In the course of comparing my individual-based model with Cristian Picioreanu's biomass-based model of biofilms, it became apparent that an objective comparison of models requires an objective, quantitative comparison of biofilm structures - apart from growth the main model predictions.
But measuring biofilm structure is not an easy task, after all, we are trying to condense the information content of a complex 3D structure into a small set of 1D shape measures. What measures should be used? Which aspect of biofilm structure is highlighted by which measure? What about interdependencies of measures? Which measures can be related to biofilm processes such as mass transfer? Which measures allow differentiation of similar biofilm structures and which measures capture the similarities? Can a set of measures be found that captures all biologically important aspects of biofilm structure?
During the last decade, a number of groups in the biofilm field have studied a variety of measures, mostly drawn from the Image Analysis literature, applied to their particular biofilms. I believe that a consensus regarding the set of suitable measures can only be reached by a comparative study of all candidate measures applied to a broad range of biofilm structures that exemplify the diversity of biofilms.
I suggested this project to a number of colleagues known for their work on biofilm structure, and we agreed to team up for this comparative study.
The first step is to compile a suitable test suite of images, and this is were your contribution would be very much appreciated.
João Xavier has set up a web site where you can submit biofilm images into a database. Soon you will be able to analyse all images in the database with João's own algorithms online. The plan is to add other published biofilm image analysis procedures to this set as the next step.